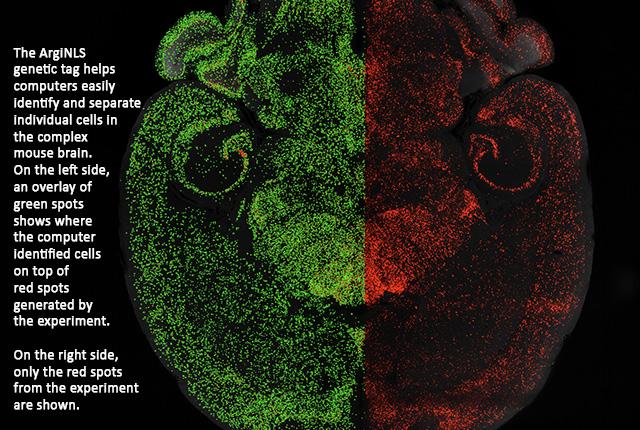
Genetic tag strategy helps automate single-cell detection
A novel method improves quantitative accuracy of single-cell brain mapping without harming neurons or disrupting behavior.Media Contact: Email mediarelations@uw.edu
An approach to genetic labeling of a cell’s nucleus with fluorescence appears to have several advantages for brain mapping at the single-cell level.
“Conventional fluorescent protein modifications used to discriminate single cells through nuclear targeting often exhibit significant signal outside of the nucleus or are detrimental to cellular health and incompatible with behavioral experiments,” said Sam Golden, describing the need for other options.
He is an assistant professor of biological structure at the the University of Washington School of Medicine in Seattle.
The method to overcome these limitations is described July 29 in PNAS, the Proceedings of the National Academy of Sciences. The lead author is Eric Szelenyi, an acting assistant professor of biological structure and a scientist in Golden’s neurosciences lab. Szelenyi headed the design of this strategy.
“These technical limitations have existed since the 1980s, and until now, have greatly impeded how scientists can count cells using fluorescence microscopy images and computers. Our approach enables more precision and enrichment of single-cell quantification — which is essential in cleanly defining how phenomenon such as complex behavior, cognition and disease states are encoded across the brain,” he said.
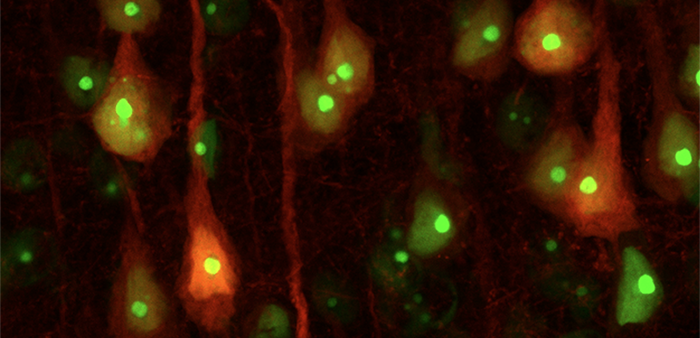
The Golden Lab is interested in the cell mechanisms and circuitry that underlie neuropsychiatric disorders such as depression, aggression and addiction. A major challenge to doing so is that brain tissues are dense and complex.
Szelenyi, Golden and their team have introduced a method to fully localize fluorescent labels into the nucleus of cells. Using a short genetic tag, they enabled more faithful, rapid detection of single cells. When used with genetically encoded fluorescent proteins, for example, the proteins glow like a lantern on a ship at night.
Among the roughly 100 million cells in a mouse brain, the method makes individual cells more noticeable, highlighting their whereabouts in intricate brain regions. The modified proteins function universally across colors such as lime green, scarlet and yellow.
The tag takes advantage of the biochemical properties of arginine, one of the basic amino acids or building blocks of proteins. The scientists formulated a specific genetic configuration that is generalizable, and named the tag “arginine-rich nuclear localization signal,” or ArgiNLS.
The researchers found that this tagging strategy did not alter the physiology of brain cells. Also, when used in mice during behavioral studies, it did not change how the mice acted. The animals continued to curiously explore their environment, to perform certain tasks and to follow their usual night and day activity cycles.
Tagging cells helps in the process of image segmentation, which makes visuals easier to interpret. Image segmentation, for example, enables driverless cars to watch the road and look out for dangers.
In the case of two- and three-dimensional brain imaging, the arginine-rich nuclear localization signal significantly improved the process of image segmentation by machine-learning, and lowered the costs. For instance, with whole-brain mapping, a machine-learning single cell classifier could be trained with comparatively fewer images and decreased computational expense.
This benefit accelerates the experimental speed required to go from complex tissue to big data.
“It enhances and joins the process where AI and ML falls short, by optimizing the biology rather than algorithms or microscope,” Szelenyi explained.
He suggested it is likely to advance the quantification of single cells in both small- and large-scale image datasets, and to enhance machine-learning capabilities in classifying cells in brain images and beyond.
The scientists noted that, although the tagging system performed effectively in liver tissue, it is unknown how well it might work in other peripheral tissues or in non-murine animal species.
The project was supported by the UW Center of Excellence in Addiction, Pain and Emotion Molecular Genetics Resource Core (P3ODA048736). The arginine-rich nuclear location signal has an associated patent with UW CoMotion, a technology-transfer service.
For details about UW Medicine, please visit https://uwmedicine.org/about.