
Biologists train AI to generate medicines and vaccines
Machine learning, used to design proteins with a variety of functions, is doing things scientists didn't realize it was capable of.Media Contact: Leila Gray, leilag@uw.edu, 206.475.9809
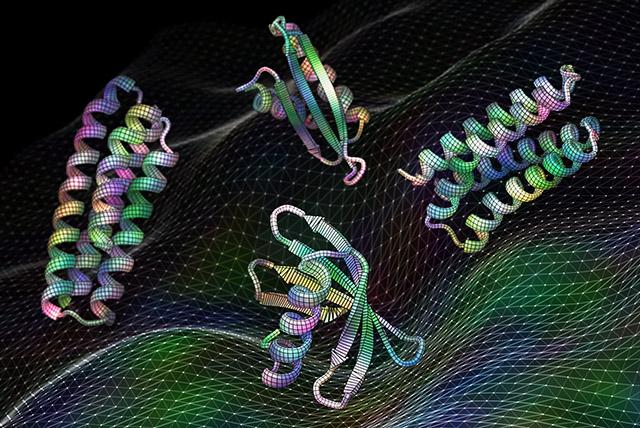
Scientists have developed artificial intelligence software that can create proteins that may be useful as vaccines, cancer treatments, or even tools for pulling carbon pollution out of the air.
This research, reported today in the journal Science, was led by the University of Washington School of Medicine and Harvard University. The article is titled "Scaffolding protein functional sites using deep learning."
“The proteins we find in nature are amazing molecules, but designed proteins can do so much more,” said senior author David Baker, an HHMI Investigator and professor of biochemistry at UW Medicine. “In this work, we show that machine learning can be used to design proteins with a wide variety of functions.”
For decades, scientists have used computers to try to engineer proteins. Some proteins, such as antibodies and synthetic binding proteins, have been adapted into medicines to combat COVID-19. Others, such as enzymes, aid in industrial manufacturing. But a single protein molecule often contains thousands of bonded atoms; even with specialized scientific software, they are difficult to study and engineer.
Inspired by how machine learning algorithms can generate stories or even images from prompts, the team set out to build similar software for designing new proteins. “The idea is the same: neural networks can be trained to see patterns in data. Once trained, you can give it a prompt and see if it can generate an elegant solution. Often the results are compelling — or even beautiful,” said lead author Joseph Watson, a postdoctoral scholar at UW Medicine.
The team trained multiple neural networks using information from the Protein Data Bank, which is a public repository of hundreds of thousands of protein structures from across all kingdoms of life. The neural networks that resulted have surprised even the scientists who created them.
The team developed two approaches for designing proteins with new functions. The first, dubbed “hallucination” is akin to DALL-E or other generative A.I. tools that produce new output based on simple prompts. The second, dubbed “inpainting,” is analogous to the autocomplete feature found in modern search bars and email clients.
“Most people can come up with new images of cats or write a paragraph from a prompt if asked, but with protein design, the human brain cannot do what computers now can,” said lead author Jue Wang, a postdoctoral scholar at UW Medicine. “Humans just cannot imagine what the solution might look like, but we have set up machines that do.”
To explain how the neural networks ‘hallucinate’ a new protein, the team compares it to how it might write a book: “You start with a random assortment of words — total gibberish. Then you impose a requirement such as that in the opening paragraph, it needs to be a dark and stormy night. Then the computer will change the words one at a time and ask itself ‘Does this make my story make more sense?’ If it does, it keeps the changes until a complete story is written,” explains Wang.
Both books and proteins can be understood as long sequences of letters. In the case of proteins, each letter corresponds to a chemical building block called an amino acid. Beginning with a random chain of amino acids, the software mutates the sequence over and over until a final sequence that encodes the desired function is generated. These final amino acid sequences encode proteins that can then be manufactured and studied in the laboratory.
The team also showed that neural networks can fill in missing pieces of a protein structure in only a few seconds. Such software could aid in the development of new medicines.
“With autocomplete, or “Protein Inpainting”, we start with the key features we want to see in a new protein, then let the software come up with the rest. Those features can be known binding motifs or even enzyme active sites,” explains Watson.
Laboratory testing revealed that many proteins generated through hallucination and inpainting functioned as intended. This included novel proteins that can bind metals as well as those that bind the anti-cancer receptor PD-1.
The new neural networks can generate several different kinds of proteins in as little as one second. Some include potential vaccines for the deadly respiratory syncytial virus, or RSV.
All vaccines work by presenting a piece of a pathogen to the immune system. Scientists often know which piece would work best, but creating a vaccine that achieves a desired molecular shape can be challenging. Using the new neural networks, the team prompted a computer to create new proteins that included the necessary pathogen fragment as part of their final structure. The software was free to create any supporting structures around the key fragment, yielding several potential vaccines with diverse molecular shapes.
When tested in the lab, the team found that known antibodies against RSV stuck to three of their hallucinated proteins. This confirms that the new proteins adopted their intended shapes and suggests they may be viable vaccine candidates that could prompt the body to generate its own highly specific antibodies. Additional testing, including in animals, is still needed.
“I started working on the vaccine stuff just as a way to test our new methods, but in the middle of working on the project, my two-year-old son got infected by RSV and spent an evening in the ER to have his lungs cleared. It made me realize that even the ‘test’ problems we were working on were actually quite meaningful,” said Wang.
“These are very powerful new approaches, but there is still much room for improvement,” said Baker, who was a recipient of the 2021 Breakthrough Prize in Life Sciences. “Designing high activity enzymes, for example, is still very challenging. But every month our methods just keep getting better! Deep learning transformed protein structure prediction in the past two years, we are now in the midst of a similar transformation of protein design.”
This project was led by Jue Wang, Doug Tischer, and Joseph L. Watson, who are postdoctoral scholars at UW Medicine, as well as Sidney Lisanza and David Juergens, who are graduate students at UW Medicine. Senior authors include Sergey Ovchinnikov, a John Harvard Distinguished Science Fellow at Harvard University, and David Baker, professor of biochemistry at UW Medicine.
Compute resources for this work were donated by Microsoft and Amazon Web Services.
Funding was provided by the Audacious Project at the Institute for Protein Design; Microsoft; Eric and Wendy Schmidt by recommendation of the Schmidt Futures; the DARPA Synergistic Discovery and Design project (HR001117S0003 contract FA8750-17-C-0219); the DARPA Harnessing Enzymatic Activity for Lifesaving Remedies project (HR001120S0052 contract HR0011-21-2-0012); the Washington Research Foundation; the Open Philanthropy Project Improving Protein Design Fund; Amgen; the Human Frontier Science Program Cross Disciplinary Fellowship (LT000395/2020-C) and EMBO Non-Stipendiary Fellowship (ALTF 1047-2019); the EMBO Fellowship (ALTF 191-2021); the European Molecular Biology Organization (ALTF 139-2018); the “la Caixa” Foundation; the National Institute of Allergy and Infectious Diseases (HHSN272201700059C), the National Institutes of Health (DP5OD026389); the National Science Foundation (MCB 2032259); the Howard Hughes Medical Institute, the National Institute on Aging (5U19AG065156); the National Cancer Institute (R01CA240339); the Swiss National Science Foundation; the Swiss National Center of Competence for Molecular Systems Engineering; the Swiss National Center of Competence in Chemical Biology; and the European Research Council (716058).
Written by Ian Haydon, UW Medicine Institute for Protein Design
For details about UW Medicine, please visit https://uwmedicine.org/about.