
Fundamental protein structure engineered in lab
The advance paves the way for building entirely novel, custom-designed antibodies, enzymes and other biologically active proteins.Media Contact: UW Medicine Media Relations: mediarelations@uw.edu, 206.543.3620
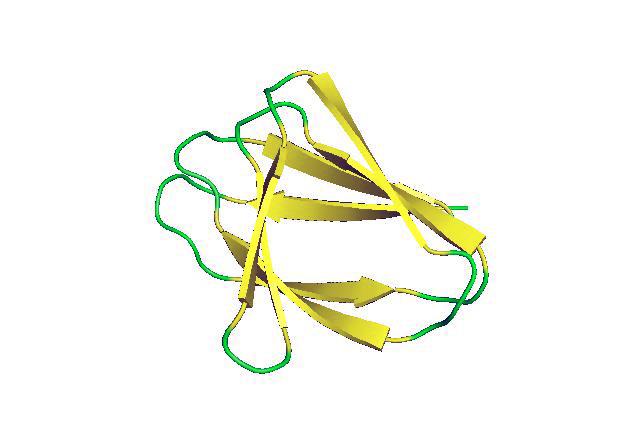
For the first time scientists have successfully designed non-local beta-sheet proteins from scratch. These structures will be essential for creating artificial antibodies and active sites for enzymes and ligand-binding proteins, which are critical to many processes in living things.
The advance opens the way for custom-designing complex proteins for therapeutics, diagnostics and industrial enzymes, said Enrique Marcos, who with Tamuka M. Chidyausiku, was a lead author of the paper reporting the work. The results are published in the Oct. 29 online issue of the scientific journal, Nature Structural & Molecular Biology.
“Non-local beta-sheets were among the most fundamental structures found in natural proteins that had not yet been designed from scratch,” Marcos said.
The research was performed at the Institute of Protein Design, directed by David Baker,

professor of biochemistry at the University of Washington School of Medicine. Marcos is a former Marie Curie postdoctoral fellow in biochemistry. He now at the Institute for Research in Biomedicine, Barcelona, Spain. Chidyausiku is a graduate student in biochemistry at the UW School of Medicine.
Proteins are made of chains of amino acids. Once synthesized, these chains spontaneously fold and twist into complex shapes that determine their function. While folding, the chains create two relatively stable types of structures, called alpha-helices and beta-sheets. To form the helices, the chain twists around hydrogen bonds across the helix that stabilize its structure. Beta-sheets, on the other hand, are created when segments of the amino acid chain line up side by side. Hydrogen bonds forming between the strands create sheet-like structures.
In previous work, published in September, another Institute for Protein Design project showed for the first time that it was possible to create barrel-like structures with local beta-sheets. In these structures, a strand of the amino acid chain runs one direction, then makes a tight looping hairpin turn to run alongside the first strand in the opposite direction. The two paired strands create the sheet by forming hydrogen bonds between each other. The chain can then make another hairpin turn and come back to add another strand to the sheet, and so on.
In many enzymes, antibodies and other key proteins, however, the formation of beta-sheets is much more complex. Instead of having the amino acid chain simply turn back on itself to create paired strands, the first strand of the beta-sheet might continue on to form another structure before returning to create the second strand of the beta-sheet. The chain may go on to create other structures before again returning to create another strand. As a result, the strands that make up the beta sheet may come from strings of amino acids that are far apart from each other along the length on the amino acid chain — that is, they are non-local. These structures, complex as they are, are the result of millions of years of evolution and are typical of proteins found in nature.
In the new study, the researchers used a computer program developed in the Baker lab, called Rosetta. The program takes advantage of the fact that the shape an amino acid chain will assume is driven by the forces of attraction and repulsion between the amino acids of the chain and the surrounding fluid. The program can calculate the shape that best balances out these forces so that the chain achieves the lowest overall energy. In so doing, the program can predict the shape a given chain of amino acids will likely take.
By studying the structure of beta-sheets that occur in nature, the researchers identified the rules governing non-local beta-sheet formation. They then used the Rosetta program to apply rules to design amino acid chains that would form non-local beta-sheets. To keep a strand from bonding to the strand immediately following it, the researchers created what resembles an arch between them that, unlike the hairpin loops, puts the two local strands at an angle that prevents them from lining up next to each other.

Using this strategy, the researchers were able to create a sandwich-like structure made of two non-local beta-sheets. It has the shape of a double-stranded beta-helix. This is more colloquially known as the jelly roll fold owing to the similarity to a jelly roll cake. Proteins created by this method should provide stable scaffolding on which active sites can be created that can mimic the binding site of antibodies, perform enzymatic functions, and bind targeted metals, compounds and proteins, said Chidyausiku.
“These proteins are also much smaller than those found in nature and easier to modify so they should be easier to make into drugs," he exaplained, "because they are much more stable, they can be used in environments too harsh for natural proteins to function or survive.”
This research was performed in collaboration with the groups at the Sgourakis lab at the University of California, Santa Cruz, and the Tripsines lab at the Masaryk University, Czech Republic.
The work was supported by the Howard Hughes Medical Institute, Open Philanthropy, Defense Threat Reduction Agency, Marie Curie International Outgoing Fellowship, Ministry of Education, Youths and Sports of the Czech Republic, Grant Agency of Masaryk University, National Institute of General Medical Sciences, and the Office of the Director of the National Institutes of Health, under a High End Instrumentation Grant.
News release written by Michael McCarthy.
For details about UW Medicine, please visit https://uwmedicine.org/about.