
Animals in motion: Toolkit tracks behavior in 3D
A set of open-source software modules could improve scientists’ ability to analyze the movement and poses of animals (and people).Media Contact: Leila Gray, 206.475.9809, leilag@uw.edu
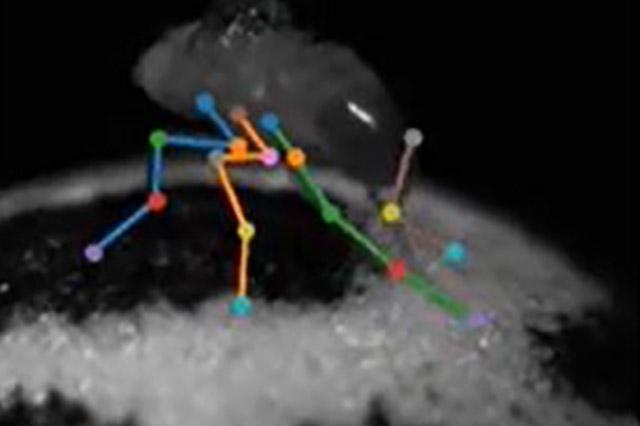
From a fly walking on a treadmill, to a mouse grasping for, and sometimes missing, a kibble, recording the ways creatures move is important in studying animal behaviors. Such observations can reveal how limbs, paws, wings, and other parts operate to carry out a variety of activities, how animals maintain balance in precarious situations, and many aspects of their locomotion and body language.
A new toolkit is now available to help scientists better analyze animals and humans in motion.
Called Anipose, a conflation of animal pose and pronounced “any pose,” the set of modules, along with directions to use them, are now available as an open source software toolkit. What makes this toolkit different is that it assists with 3D analytics. This makes it more closely aligned with the shape of animals and the space they inhabit, compared to flat, two dimensional recordings. It was built based on a popular 2D tracking method called DeepLabCut.
“Advances in computer vision and artificial intelligence enable markerless tracking from 2D video,” the Anipose developers noted, “but most animals live and move in 3D.” They also explained that markers are often impractical when studying natural behaviors in an animal’s environment, tracking several different body parts at once, or observing small animals.
The development and testing of the program was headed by the lab of John Tuthill, assistant professor of physiology and biophysics at the University of Washington School of Medicine, and Bing Brunton, associate professor of biology in the UW College of Arts & Sciences. Tuthill and his team are known for their explorations of several aspects of animal movement, such as how bodily sensation provides cues for navigation. The Brunton Lab uses big data analysis and modeling to investigate the brain and behavior.
One of the demonstration videos shows how Anipose can capture the six-legged walking of a fly on a treadmill by recording with cameras from several directions. That study allowed the scientists to determine the role of joint rotation of the insect’s middle legs, and the ways it differs from the flexing of the front and back legs, in how the fly controls walking. Another video, a rendition of a person’s palm and fingers hovering and turning above a table, results in a beautiful, ballet-like figure floating in white space.
The developers hope that Anipose eventually may find many uses, not only in animal behavior studies, but also in medicine and other fields.
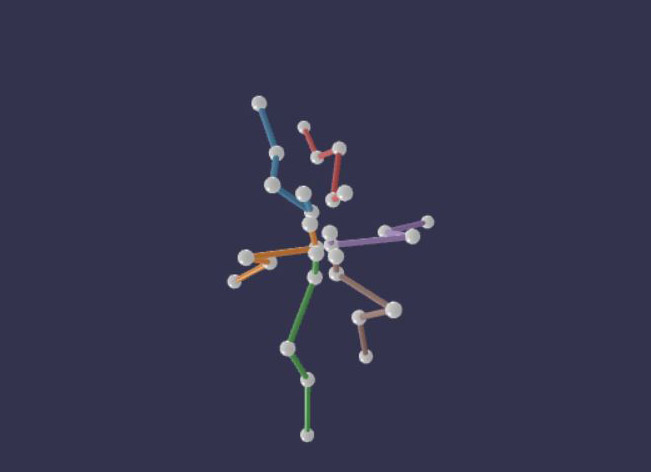
The scientists hope that 3D markerless solutions will make quantitative movement analysis more accessible to basic scientists and to clinicians. For example, such solutions might make it less costly and more convenient to evaluate motor disorders and to assess recovery after treatment.
“Many neurological disorders, including some commonly thought of as cognitive disorders, affect walking gait and coordination of the arms and hands,” the scientists explained. Anipose could become an aid in the diagnosis, assessment, and rehabilitative treatment of movement and neurodegenerative disorders.
It is also imaginable that such a toolkit could eventually assist in coaching dancers, sports athletes and actors. It has the advantage of not requiring them them to wear markers or a special garment for analyzing their movements or body positioning in 3D.
The toolkit, and tests of abilities for studies of mice, humans, and flies in motion, are described this week in Cell Reports, one of the scientific journals of Cell Press. The paper is titled “Anipose: a toolkit for robust markerless 3D pose estimation.”
Directions for installing Anipose and a tutorial are available at:
Anipose — Anipose 0.8.1 documentation
The code and documentation were developed by Pierre Karashchuk and Katie Rupp. Testing datasets for the fly were done by Evyn S. Dickinson at the UW, and for the mouse by Elisha Sanders and Eiman Azim at the Salk Institute. Additional contributions were made by Julian Pitney. John Tuthill, and Bingni W. Brunton mentored the toolkit creators.
The project was supported by a National Science Foundation Research Fellowship, fellowships from the UW Institute for Neuroengineering and Center for Neurotechnology, National Institutes of Health (F31NS115477, NS088193, DP2NS105555, R01NS111479, R01NS102333 and U19NS104655 and U19NS112959), the Searle Scholars Program, The Pew Charitable Trusts, the McKnight Foundation, Sloan Research Fellowship, and the Washington Research Foundation.
For details about UW Medicine, please visit https://uwmedicine.org/about.